Reactive execution¶
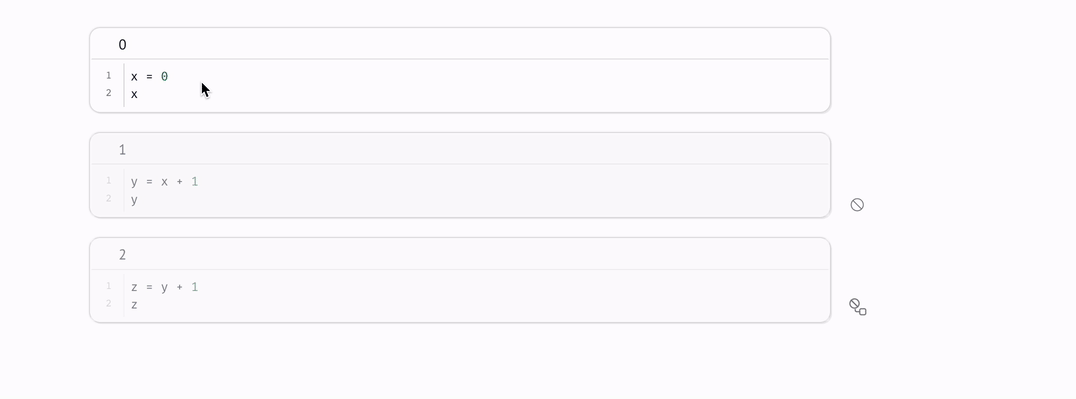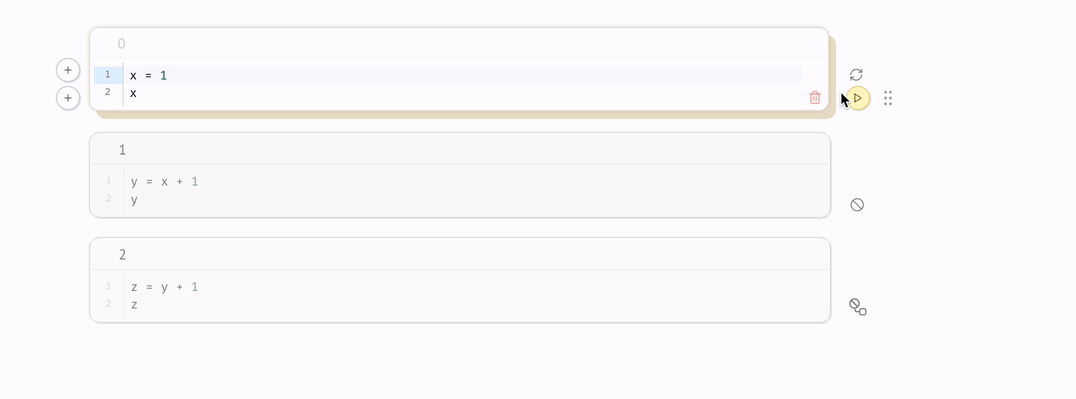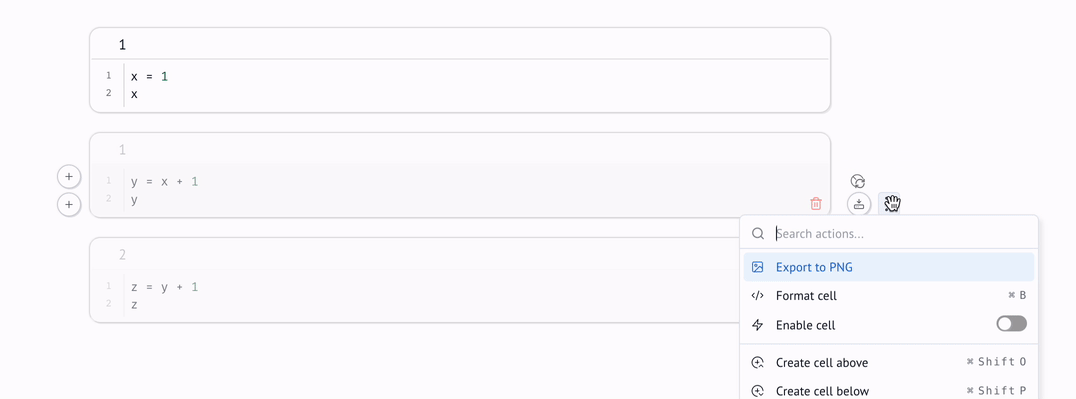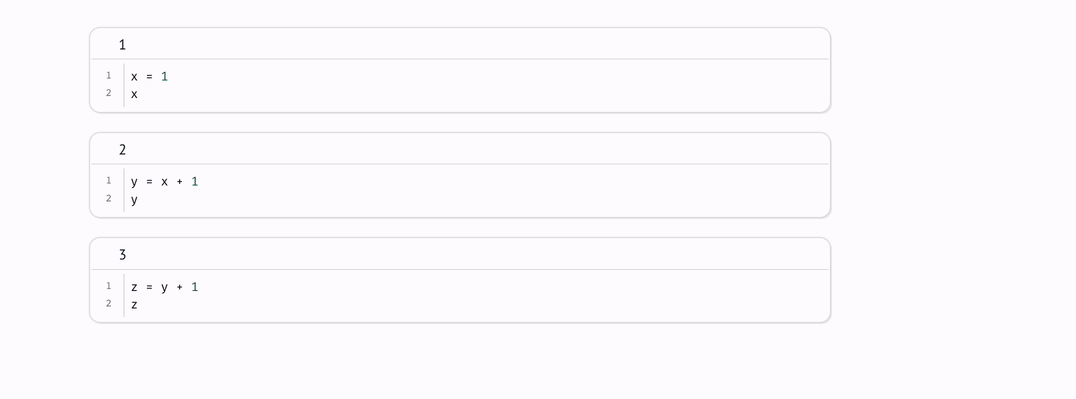
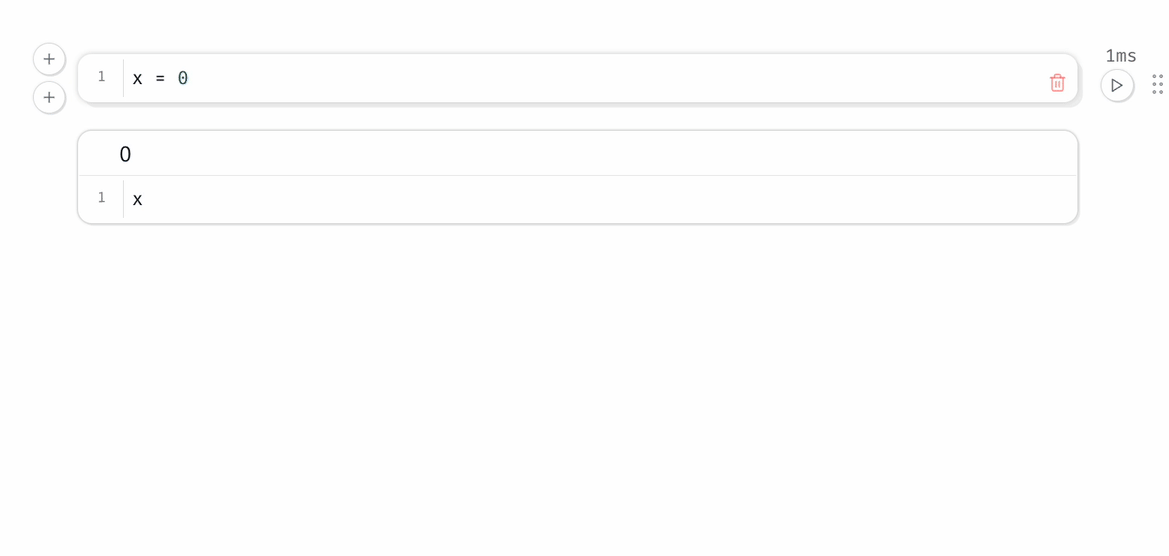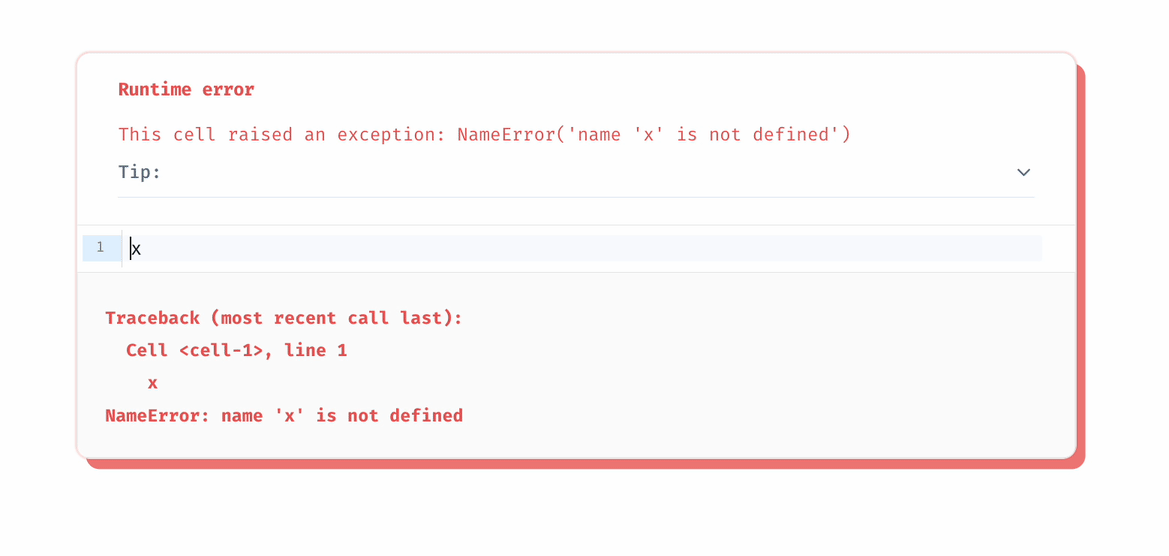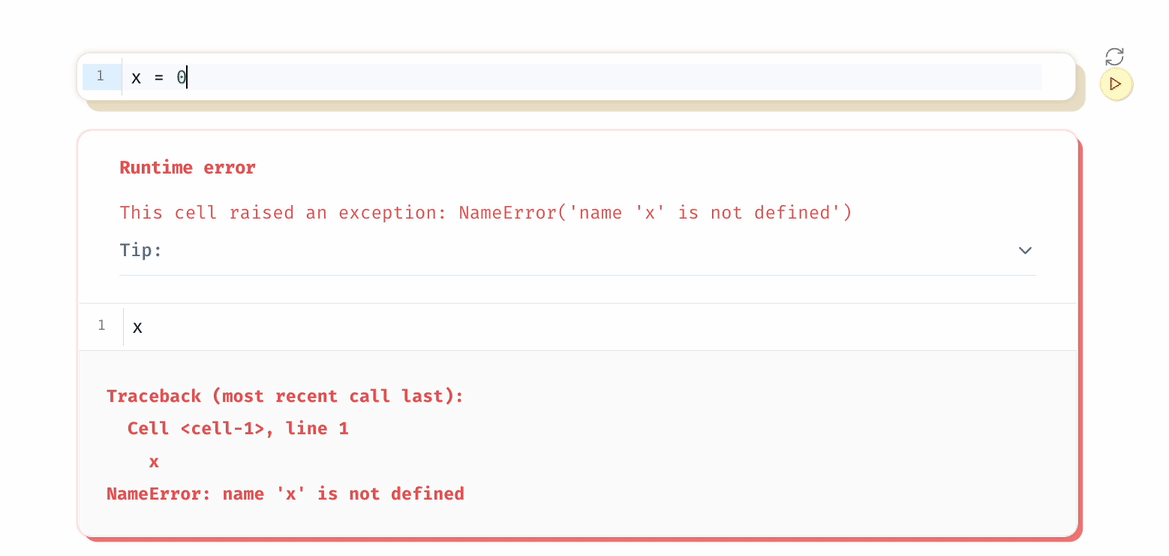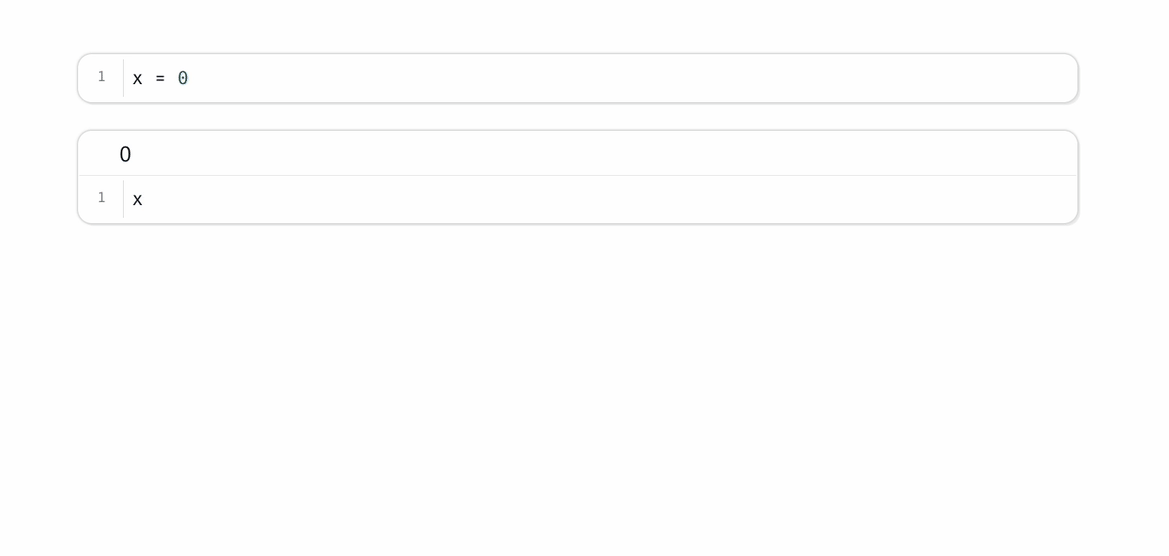
Every marimo notebook is a directed acyclic graph (DAG) that models how data flows across blocks of Python code, i.e., cells.
marimo react to code changes, automatically executing cells with the latest data. Execution order is determined by the DAG, not by the order of cells on the page.
Reactive execution is based on a single rule:
Runtime Rule
When a cell is run, marimo automatically runs all other cells that reference any of the global variables it defines.
Working with expensive notebooks
marimo gives you tools that make it easy to work with expensive notebooks. For example, the runtime can be configured to be lazy, only running cells when you ask for them to be run and marking affected cells as stale instead of auto-running them. See our guide on working with expensive notebooks for more tips.
References and definitions¶
A marimo notebook is a DAG where nodes are cells and edges are data dependencies. marimo creates this graph by statically analyzing each cell (i.e., without running it) to determine its
references, the global variables it reads but doesn’t define;
definitions, the global variables it defines.
Global variables
A variable can refer to any Python object. Functions, classes, and imported names are all variables.
There is an edge from one cell to another if the latter cell references any global variables defined by the former cell. The rule for reactive execution can be restated in terms of the graph: when a cell is run, its descendants are run automatically.
Global variable names must be unique¶
To make sure your notebook is DAG, marimo requires that every global variable be defined by only one cell.
Local variables
Variables prefixed with an underscore are local to a cell (.e.g., _x). You
can use this in a pinch to fix multiple definition errors, but try instead to
refactor your code.
This rule encourages you to keep the number of global variables in your program small, which is generally considered good practice.
Local variables¶
Global variables prefixed with an underscore (e.g., _x) are “local” to a
cell: they can’t be read by other cells. Multiple cells can reuse the same
local variables names.
If you encapsulate your code using functions and classes when needed, you won’t need to use many local variables, if any.
Avoid mutating variables¶
marimo’s reactive execution is based only on the global variables a cell reads and the global variables it defines. In particular, marimo does not track mutations to objects, i.e., mutations don’t trigger reactive re-runs of other cells. It also does not track the definition or mutation of object attributes. For this reason, avoid defining a variable in one cell and mutating it in another.
If you need to mutate a variable (such as adding a new column to a dataframe), you should perform the mutation in the same cell as the one that defines it, Or try creating a new variable instead.
Examples¶
Create a new variable instead of mutating an existing one.
Don’t do this:
l = [1]
l.append(2)
Instead, do this:
l = [1]
extended_list = l + [2]
Mutate variables in the cells that define them.
Don’t do this:
df = pd.DataFrame({"my_column": [1, 2]})
df["another_column"] = [3, 4]
Instead, do this:
df = pd.DataFrame({"my_column": [1, 2]})
df["another_column"] = [3, 4]
Why not track mutations?
Tracking mutations reliably is a fundamentally impossible task in Python; marimo could never detect all mutations, and even if we could, reacting to mutations could result in surprising re-runs of notebook cells. The simplicity of marimo’s static analysis approach, based only on variable definitions and references, makes marimo easy to understand and encourages well-organized notebook code.
Runtime configuration¶
Through the notebook settings menu, you can configure how and when marimo runs cells. In particular, you can disable autorun on startup, disable autorun on cell execution, and enable a powerful module autoreloader. Read our runtime configuration guide to learn more.
Disabling cells¶
Sometimes, you may want to edit one part of a notebook without triggering automatic execution of its dependent cells. For example, the dependent cells may take a long time to execute, and you only want to iterate on the first part of a multi-cell computation.
For cases like this, marimo lets you disable cells: when a cell is disabled, it and its dependents are blocked from running.
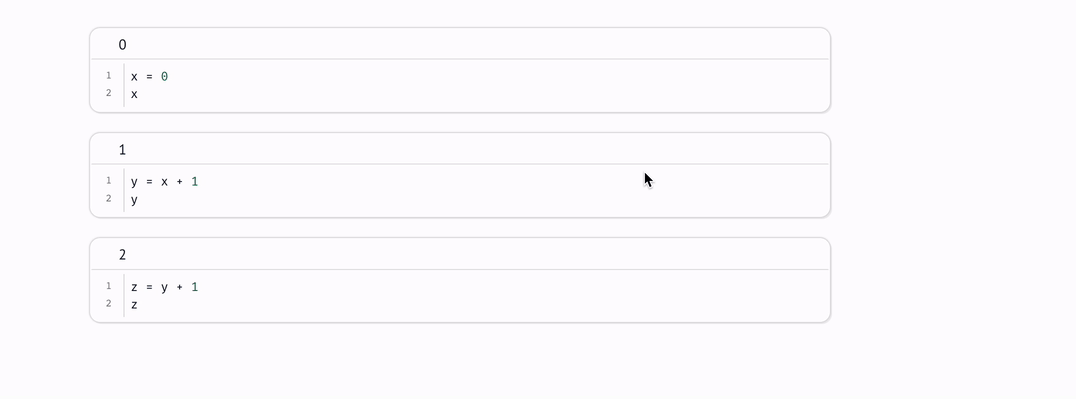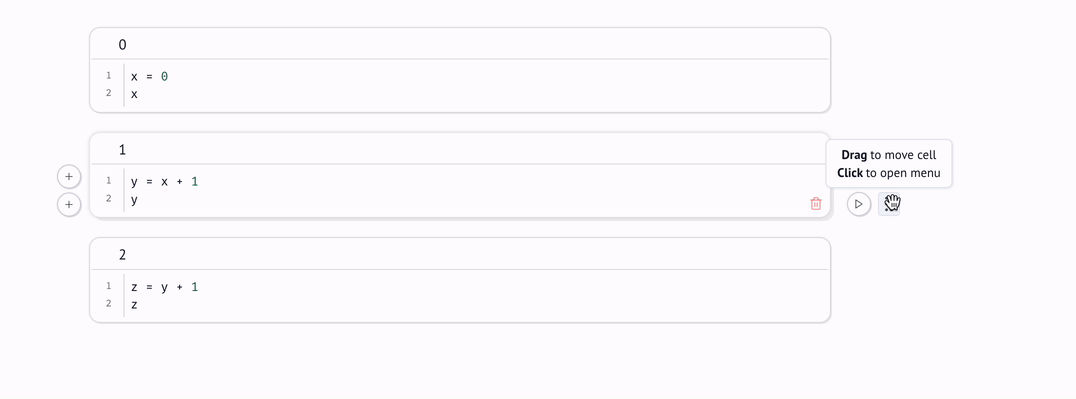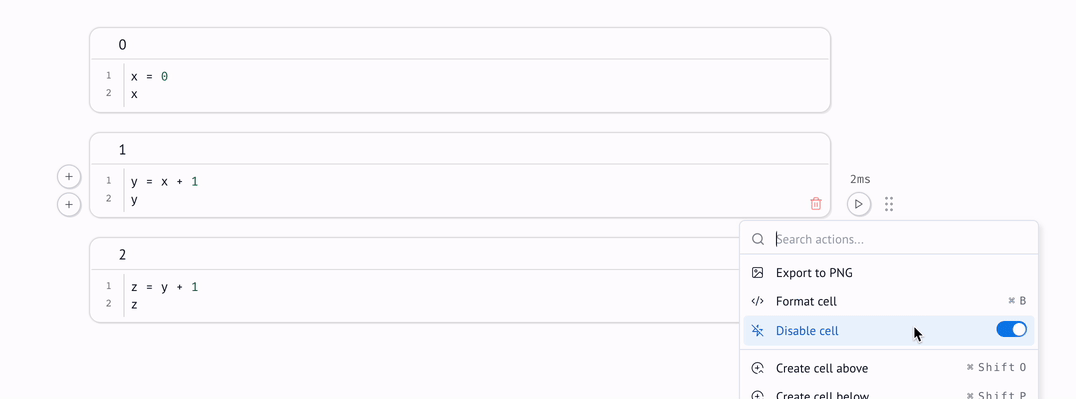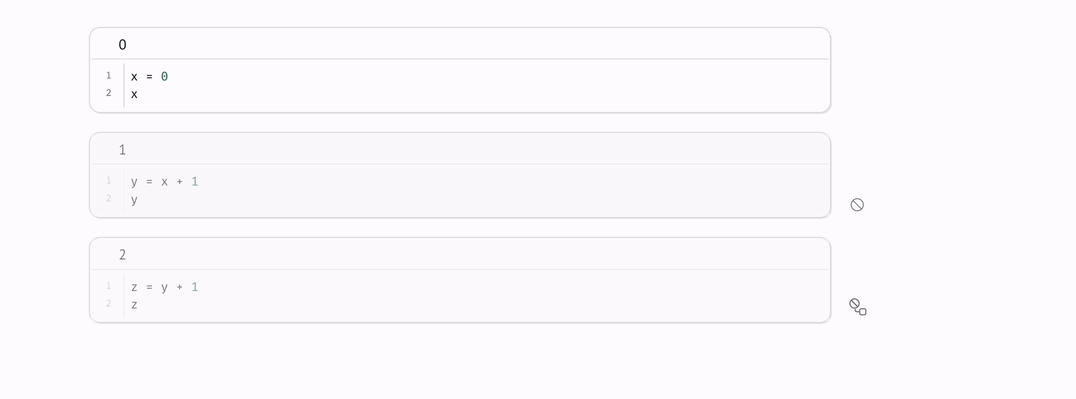
When you re-enable a cell, if any of the cell’s ancestors ran while it was disabled, marimo will automatically run it.